Chapter 7: Architecture of the Program¶
Software Architecture¶
In practice, computational materials science research encompasses three distinctive steps, viz. (1), constructing the geometrical data or structure of the system (2), performing computations and, (3), obtaining results by the post processing of calculated data. All these steps can be effectively coupled by means of modeling platform such that a user can create, visualize, and share various input data files and run it with appropriate programs to obtain output data to analyze the results.
By integrating molecular (or crystal) visualizer, scripting tools (in this case, Python and Jmol scripting) and other standard features of an IDE (integrated development environment), a user can interactively build or manipulate structures of materials or molecules, and its atomistic properties can be calculated with appropriate ab initio programs from either a local or a remote machine. A number of built-in tools, scripts are provided for the analysis purpose. To demonstrate the platform we have used DFTB+ as well as Siesta electronic structure code along with ASE (atomic simulation environment) and OpenMD molecular dynamics package.
The programming language, Java (version 8) was chosen for this project since it is free, strongly object-oriented and shows its neutrality to the various operating systems. The object oriented programming is arguably the most popular software development technique which effectively manages the complexity in code development [9]. We preferred Java, since (1) it does not use pointers, and (2), it supports threads implicitly. Although pointers provide direct access to the computer memory, careless use of the pointers will easily leads to segmentation fault and other vulnerables. It is our personal opinion that the bugs emerged from the improper application of pointers are difficult to trace or debug, in addition to this, code with pointers is difficult to translate into other computer languages without the pointer features. Concerning threads, in this application, we used a number of threads (i.e. user threads), apart from the JVM generated daemon threads. Since the threads are inherently supported by the language one can readily create a thread by extending a Thread class. Exception handling of Java is equipped with two types, namely, unchecked and checked exception handling. And in most of the cases we used checked exceptions, as it is less tedious to implement.
The core of the DJMol application consists of (Apache-) Netbeans Platform and it can be regarded as the engine behind the Netbeans IDE. In other words, many of the technical features of DJMol program is inherited from Netbeans IDE which is initially designed for developing Java/Javascript and C/C++ applications and consists a number of utilities to increase the productivity of a user (for example, it has an advanced sourcecode editor with code completion utilities, tools for refactoring, version control systems, Git based collaboration tools etc.) and these utilities can also be used efficiently for various modeling or scripting tasks. Moreover, this platform consists of a set of independent modular software components called modules (e.g., an SSH module to communicate with an external cloud platform). Apart from this, the program supports plug-ins so that one can selectively add or remove new features into it (e.g. Python interpreter) without being re-compiled the code. Optionally, users of the code can make their own plug-ins, for example, to incorporate another ab initio package. See the below for the schematic structure of the program. To the best of our knowledge, DJMol is the first opensource modeling platform which is built from a programming IDE.
To display the molecular, crystal, nanostructures a Java library, Jmol, has been embedded into the program. The Jmol program is an open source, cross-platform and a highly independent (i.e. it does not depend on any third party libraries like Java3D or OpenGL) 3D visualization application. Apart from this, there are two distinct features associated with Jmol: (1) Jmol can be programmatically embedded into any Java code which uses Swing application programming interface (2), It supports an internal command scripting, so that a user can control or manipulate the display or send and retrieve parameters, data, commands etc. For example, to manipulate Gaussian cube files a user can effectively apply this internal scripting ability of Jmol. Unlike most of the visualizers, Jmol is capable of perceiving the molecular structure in three dimensions by applying stereographic projections. A viewer, with appropriate anaglyph spectacles, 3D perspective images of the molecules can be interactively visualized. Apart from this, Jmol consists of an UFF (uniform force field) method and it can be used to pre-optimize a variety of organic as well as inorganic molecules. To create 2D data plots we have used either Matplotlib based scripts or Jfreechart libraries. In the case of 3D data plots, (for example, the potential energy surfaces or orbital contour diagrams), a Java based opensource library jzy3d has been used.
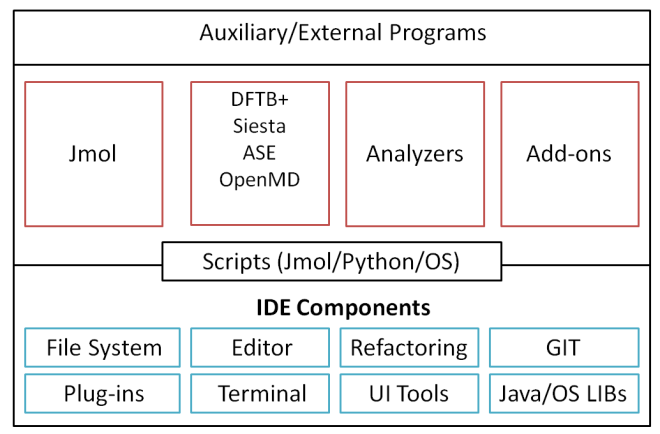
A schematic diagram of DJMol software architecture. The core of the program is an IDE which is connected to other modules or programs.¶
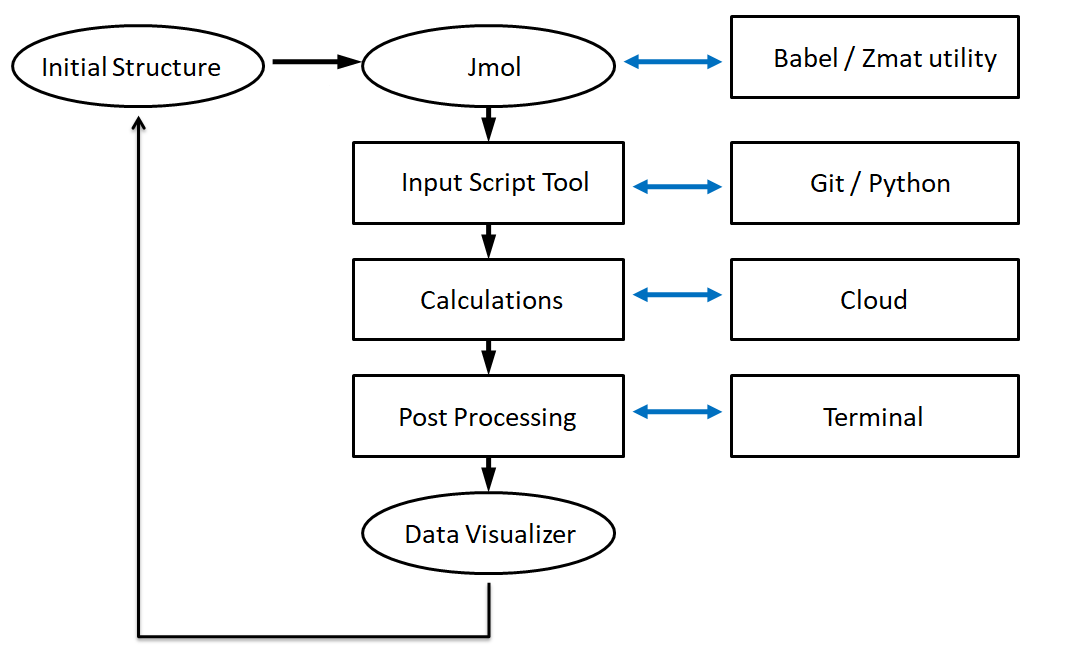
A standard workflow for a modeling task under the DJMol program (mandate workflows are indicated by black arrows).¶
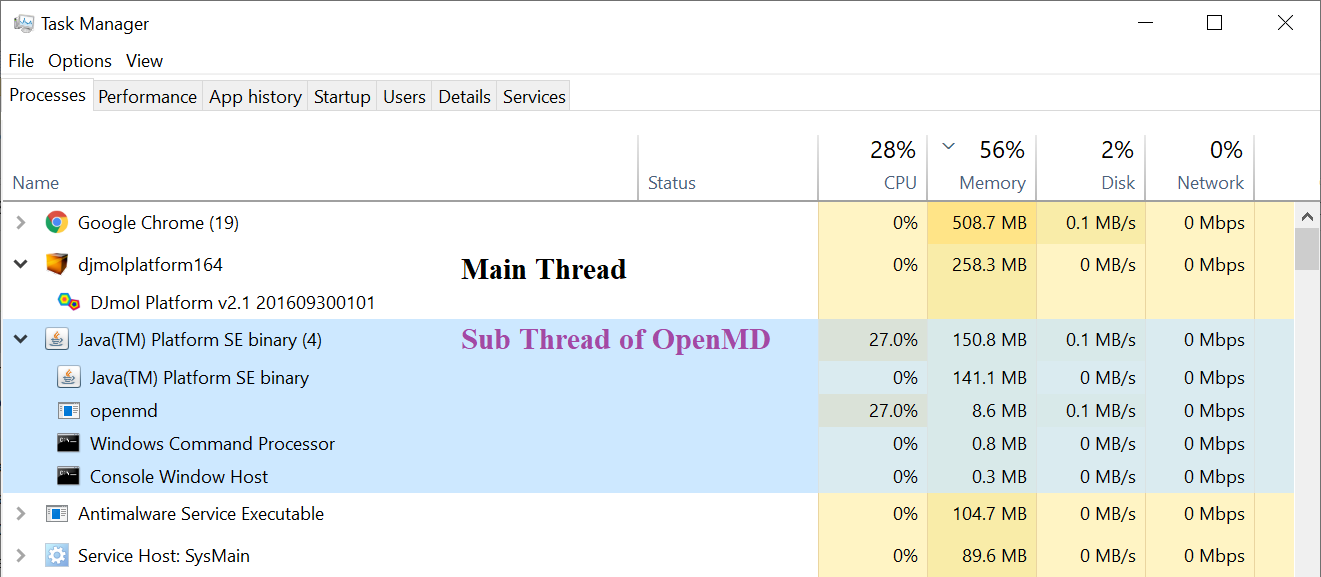
Java’s native thread is used to initiate add-ons so that its console can be controlled independently, if needed. Below figure shows Windos’ Task managers snapshot of OpenMD add-on, while it is under running (and it indicate two independent processes).
Setting up a Pyt¶